
Judy Russell, The Legal Genealogist, has provided a nice list of links to introductory information. See “DNA Basics for a Sound Foundation.”
Suffice it to say, there are basic building blocks of DNA that are represented by the letters A, C, G, and T. Our chromosomes are composed of long strings of these—3 billion, in fact. Almost all the letters are the same in every single person on the planet. Julie said that only about 10 million are different among different individuals and populations. A DNA test looks at about 700,000 of them. A location in the string of letters where the letters differ between individuals is called a SNP (pronounced “snip”). A group of inherited letters is called a haplotype.
Julie studies SNPs and haplotypes in the context of human populations. “Patterns of SNPs and haplotypes among human populations are driven by history,” she said. “As humans migrate, they bring their DNA with them.” She explained the founder effect: Not everyone in a population has the same SNPs and haplotypes. If a small number of people migrate somewhere, their most common SNPs and haplotypes are likely to be different than the parent population. They have founded a population with a different profile than the parent population. A related phenomena is isolation. If I understand correctly, newborns in an isolated population are statistically more likely to have the most common SNPs and haplotypes of their population. These effects make different populations look different genetically.
AncestryDNA uses the SNPs and haplotypes to determine three things.
- Tiny amounts of the haplotypes and SNPs associated with a population from the distant past (hundreds of thousands of years) survive in our DNA. AncestryDNA uses this information to provide your ethnicity estimates. To determine what SNPs and haplotypes are associated with distant populations, AncestryDNA uses reference panels. These are individuals whose haplotypes and SNPs are thought to be representative of the distant populations. AncestryDNA has 26 reference panels. Founder effect and isolation make ethnicity estimates easy. Migration makes ethnicity estimates difficult.
- Large amounts of shared haplotypes between two persons indicate recent common ancestors. The more closely related, the more DNA is shared. AncestryDNA uses this information to provide your DNA matches. There are several challenges in determining DNA matches. Just sharing DNA doesn’t mean you are closely related. DNA you share for other reasons is called identical by state (IBS). DNA shared because of recent common ancestry is called identical by descent (IBD). AncestryDNA has to determine the difference. Another challenge arises from the way DNA is processed in the laboratory. For any given SNP, the data coming from the lab does not differentiate between the value contributed by your father and the value coming from your mother. AncestryDNA uses tools to estimate which came from which. She didn’t say this, but I would guess that if they ever get it wrong, you could be shown relatives who aren’t really your relatives.
- In between the two extremes, AncestryDNA searches for groups of people who share large numbers of matches to others within a group. They use this information to provide your Genetic Communities.
It is possible to share no DNA at all with cousins. The closer the cousin, the higher the probability of shared DNA. Julie showed these numbers:
| Cousin | Probability of shared DNA |
| 1st | 100 |
| 2nd | 100 |
| 3rd | 98 |
| 4th | 71 |
| 5th | 32 |
| 6th | 11 |
| 7th | 3.2 |
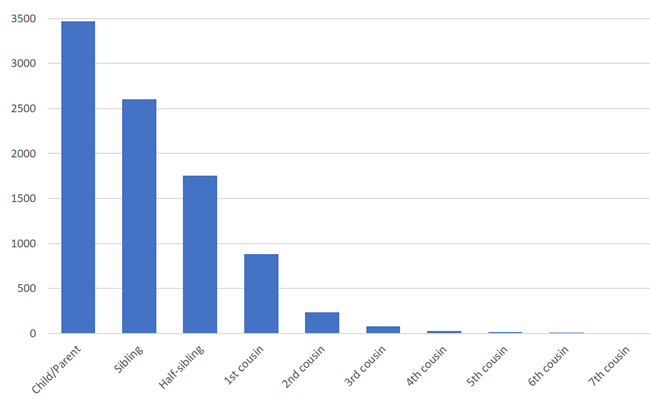
She showed a chart that looked like the one below. I think it indicated the average amount of shared DNA between two close relatives. It went by so fast, I am not certain. However, Blaine T. Bettinger provides similar data, which I’ve charted below.
Source: Blaine T. Bettinger, “The Shared CM Project – Version 2.0 (June 25, 2016),” The Genetic Genealogist (http://www.thegeneticgenealogist.com : updated 31 July 2016).
AncestryDNA uses these numbers to estimate your relationship to your DNA matches.
She covered more, but that’s about all I have time and space for here. I’m sorry that I’m not as clear as she was, but hopefully you learned something.
Chromosome inheritance diagram credit: Catherine A. Ball, et. al., “DNA Circles White Paper,” Ancestry (http://www.ancestry.com/cs/dna-help/circles/whitepaper : updated 18 November 2014), figure 2.1.
Conference audio available for purchase at http://www.playbackngs.com/7770-f342?html5player=true
ReplyDelete